Equilibrium self-assembly of anisotropic particles
Proteins are complex molecules with highly anisotropic interactions. Yet, in many pathologies, they tend to aggregate into fibers, independently of their specific physico-chemical properties.
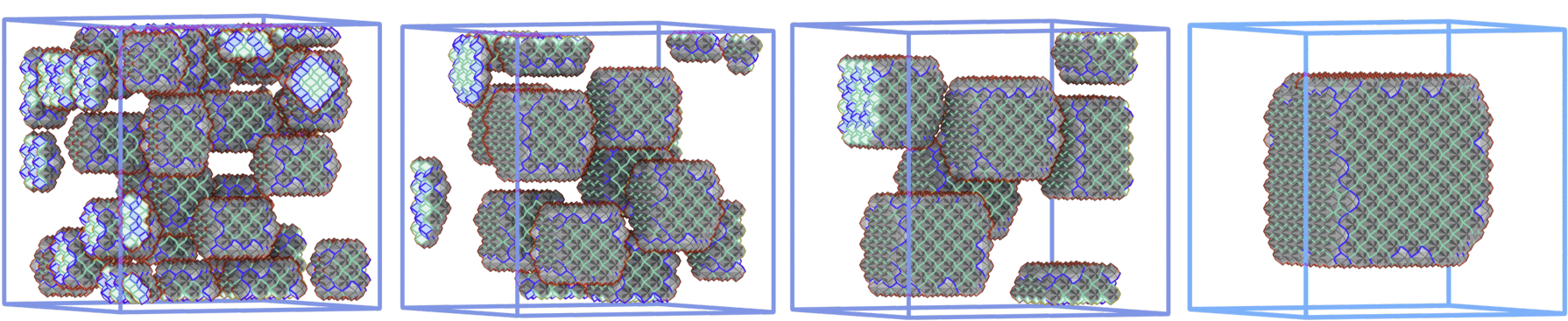
To model the self-assembly of such complex objects, I model them using three-dimensional lattice particles with anisotropic interactions. I then use Monte-Carlo simulations to understand how these particles aggregate.
Crucially, while in many cases complex interactions lead to geometrical frustration and fiber formation, the observed fibers tend te be metastable states. By performing parallel-tempering Monte Carlo simulations on symmetric superlattices, I am attempting to characterize the large-scale aggregates in which a given particle can assemble into, and especially their relative stability and nucleation times.
Another part of my current research focuses on using anisotropic interactions to control the size and shape of aggregates in equilibrium self-assembly. Using simulated annealing Monte-Carlo, I have found a design for three-dimensional particles which allows them to self-assemble into structures of different sizes by tuning a single parameter.
Self-organization of catalytically active mixtures
[TODO]
These systems develop non-reciprocal interactions which can drive their self-organization into biomolecular-condensate-like clusters. I especially focused on cases where active particles participate in biochemical reaction networks, which increases the complexity of their interactions and allows them to develop more complex behaviors.